Graphic abstract. Credit: Cell genomics (2024). DOI: 10.1016/j.xgen.2024.100537
Graduate student Hillary Layden studies transcriptional control in cancer in the lab of Scott Hiebert, the Hortense B. Ingram Chair in Cancer Research and professor of biochemistry.
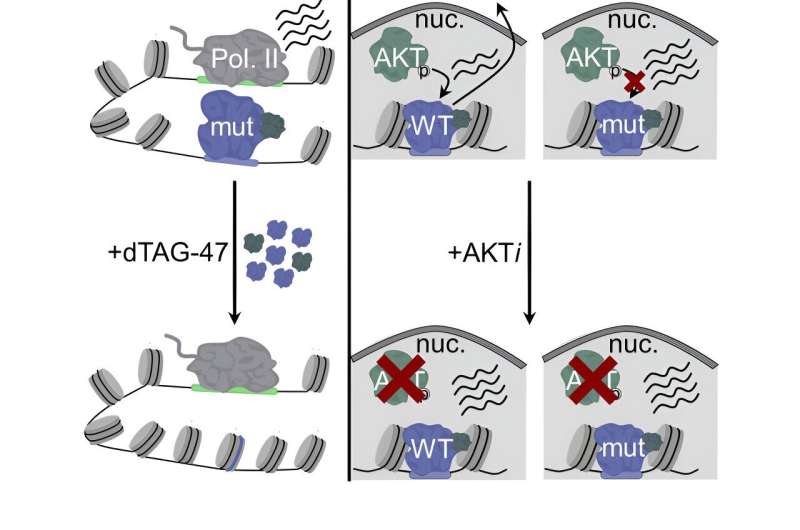
Below, Layden shares results from her research in which she used deep genomic analysis to determine how protein mutations affect gene expression to drive cancer progression.
The study is published in the journal Cell genomics.
What problem does your research address?
Mutations in proteins that control gene expression are common in all human cancers, but we do not understand how these mutations affect gene expression, contribute to disease, or affect response to treatments. In this study, we focused on identifying direct gene targets of the FOXO1 transcription factor and how mutations in FOXO1 found in B-cell lymphoma patients affect the expression of these genes.
Our approach took advantage of several recently developed technologies, including degron tags and nascent transcript analysis, to analyze transcription immediately after FOXO1 loss.
What were your findings?
We found that wild-type FOXO1 activates the transcription of genes that are key regulators of B-cell identity and are known oncogenes, while maintaining chromatin accessibility to enhancer elements. Mutant FOXO1 controls transcription in the same manner but is insensitive to signaling events that inactivate wild-type FOXO1. This allows mutant FOXO1 to maintain expression of genes that contribute to lymphoma cell growth under cellular conditions that wild-type FOXO1 cannot.
In other words, mutations in FOXO1 affect an on/off switch, locking FOXO1 in the “on” position. When FOXO1 is “on”, it increases the expression of target genes, which are critical for cell growth and survival.
What do you hope the results of this study will be?
Patients with mutations in FOXO1 are more likely to relapse under current standard of care therapy. We hope that a better understanding of how these mutations affect FOXO1 function can translate into new treatments that better serve this patient population in the long term.
In the short term, this work provides several target genes that can be used as proxies for the transcriptional function of FOXO1 in B cells. These targets can be used to identify drugs that affect the transcriptional activity of FOXO1 or other proteins required for transcriptional function of FOXO1. This work also provides much-needed clarity on how lymphoma-associated mutations in FOXO1 affect the transcription of target genes.
More information:
Hillary M. Layden et al, Mutant FOXO1 controls an oncogenic network through enhancer access, Cell genomics (2024). DOI: 10.1016/j.xgen.2024.100537
Provided by Vanderbilt University
citation: Q&A: Understanding protein mutations that affect gene expression to drive cancer progression (2024, May 30) retrieved on May 30, 2024 from https://medicalxpress.com/news/2024-05-qa- protein-mutations-affect-gene.html
This document is subject to copyright. Except for any fair agreement for study or private research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.
#Understanding #protein #mutations #affect #gene #expression #drive #cancer #progression
Image Source : medicalxpress.com
